A.I.Segmentation
OsiriX plugin for medical segmentation by A.I.
| Version 3.4.0 |
Nothing Like This
A.I.Segmentation (AIS) is an OsiriX plugin to perform segmentation of organs, tissues, or structures from medical DICOM images. The computation is based on trained neural network models, called “A.I. cores”.
AIS uses Apple Core ML and Metal technologies, which enables incredible speed and performance.
A.I. core is a Core ML model. We provide AIS Training Codeset in which you can create your own A.I. core for your target organs by yourself.
Advanced Technologies
- Optimized performance for Apple Silicon Mac *
- Apple Core ML / Metal APIs for high-performance neural network computation.
- OpenCV library is imported for various pre-segmentation image filtering.
- Dual A.I. system for complementary merge of segmentation results from two AIs.
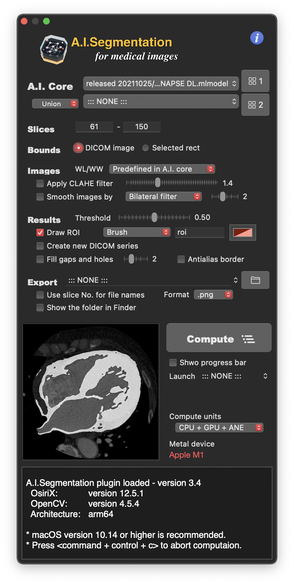
- Segmentation in OsiriX window as brush ROI or polyline.
- Direct creation of new DICOM series, useful for 3D surface rendering and STL data creation for 3D models.
- Custom neural network layers are implemented, e.g. Synaptic Transmission Regulator, Swish / Parametric Swish, GELU, Bilinear Upsampling, and so on.
* See for details: Benchmark test of M1 SoC [Article in Medium]
Rapid construction of 3D STL data of a 3D heart model.
Benchmark test on MacBook Air M1
Download
Optimized for Apple Silicon
Or, Mac with GPU recommended
Computes in macOS 10.13 or higher
* The source code of ver.1.4.1 is available from GitHub/AISegmentation_v141
|
NOTE: A.I. cores for segmentation are NOT included in A.I.Segmentation plugin itself. Please download them separately from the following ‘A.I. cores' links. At first launch, select the folder containing cores, then the cores will appear in the popup menu in AIS. |
- How to Download and Install -

Click the orange button ‘A.I.Segmentation ver...’ above.
The download site (Onedrive directory) will appear. Then click ‘Download’ in it.
A disk image (.dmg file) will be saved in your download folder. You can see the plugin file ‘A.I.Segmentation.osirixplugin’ in the disk image.

When you double click the plugin file, OsiriX will launch asking you whether the plugin should be installed into OsiriX or not. Push OK.

When you have some troubles, please check Plugin Manager, in which you can see plugins’ states, from Plugins menu in OsiriX.
|
Tips for Better Segmentation The latest A.I. core is not always the best for your images. Segmentation performance is dependent on image properties, e.g. contrast, brightness and/or object shapes, etc. In addition, because the A.I. cores for each target structure were trained in different manners, they will show different results. When you have some unexpected segmentations, change image contrast (window level/width) and try the other cores to find the best result for your cases. More, 'Dual A.I. system' in version 2.0- enables better performance by combining two cores. |
|
CARDIAC STRUCTURE |
CoreML models specialized for segmentation of cardiac structures in ECG-gated CT images. The segmentation includes the left and right ventricles, aortic/mitral valves, papillary muscles, tendons and aortic root, etc.
Image modality: ECG-gated contrast CT scan *diastole
Image plane: Normal axial view
Training dataset: Human cases
Training code: Keras and/or TensorFlow 2.5
|
CV-net SYNAPSE |
Image size : 512x512 pixel
|
U-net |
Image size : 200x200 pixel
| A.I. core - cardiac structure | February 10, 2020 |
| A.I. core - cardiac structure | February 10, 2020 |
Knowledge Source: The original neural network code of U-net was from chuckyee in GitHub.
| A.I. core - cardiac structure | Oct 25, 2021 |
| A.I. core - cardiac structure | Oct 25, 2021 |
Knowledge Source: The original neural network code of DeepLab v3 plus was from Emil Zakirov in GitHub.
|
AORTA - lumen |
Core ML models for segmentation of lumen of the aorta in CT images.
Image modality: Contrast CT scan
Image plane: Normal axial view
Training dataset: 29 human cases
Training code: TensorFlow 2.5
|
CV-net SYNAPSE |
Image size : 512x512 pixel
|
AORTA - wall |
Core ML models for segmentation of the aortic wall in CT images.
Image modality: Contrast CT scan
Image plane: Normal axial view
Training dataset: * human cases
Training code: TensorFlow 2.5
|
CALCIFICATION OF AORTIC VALVE |
CoreML models specialized for segmentation of calcification on cusps of aortic stenosis valves in ECG-gated CT images.
Image modality: ECG-gated contrast CT *diastole
Image plane: MPR view for aortic valve
Training dataset: Human cases including bicuspid aortic valves
Training code: Keras+TensorFlow (Python)
Source Code

The Xcode project of AIS plugin is available from the following GitHub site with MIT license: GitHub/AISegmentation_v141
NOTE: Keras codes or CoreML models are available from the following 'Train your original A.I. cores'.
Train Your Original A.I. Cores

Python code to train a neural network for your own A.I. cores. Available from my GitHub site: GitHub/AIS_Training_Codeset
- Main python code to manage training process
- Neural network models written in Keras functional API
- Data generator for square grayscale images
- Keras to CoreML converter for custom layers of Keras
- Other useful code examples...
|
Tips: How to look inside the NN structure? You can look inside neural network models by Netron. Have fun and challenge deep learning for your own A.I.core! |